
Rice Science ›› 2020, Vol. 27 ›› Issue (6): 445-448.DOI: 10.1016/j.rsci.2020.09.001
• • 下一篇
收稿日期:2019-12-25
接受日期:2020-04-27
出版日期:2020-11-28
发布日期:2020-11-28
. [J]. Rice Science, 2020, 27(6): 445-448.
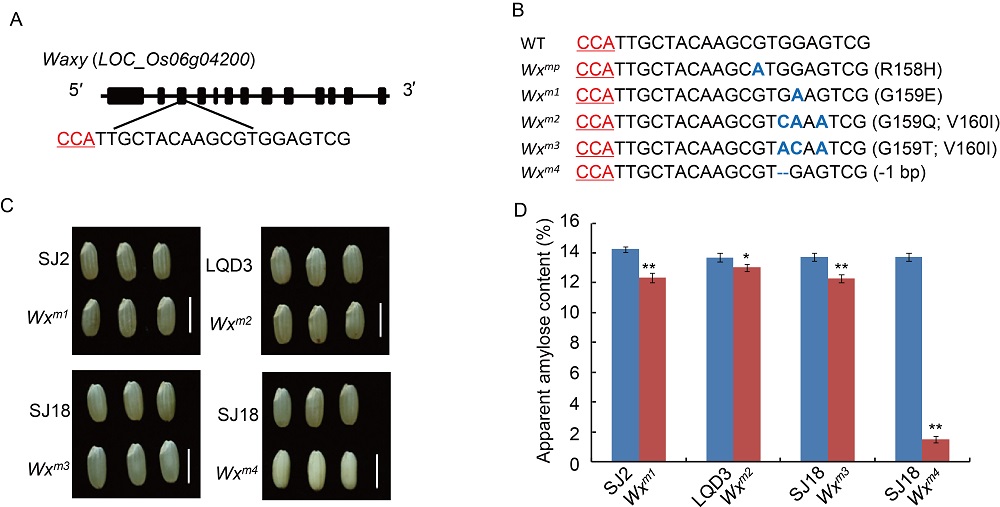
Fig. 1. Generation of lines with decreased apparent amylose content via CRISPR/Cas9-mediated base editing of Waxy gene in elite rice varieties.A, Schematic diagram of the targeted site in the Waxy gene. Black boxes and line indicate the exon and intron, respectively. The sequence of the targeted site is shown with the protospacer adjacent motif (PAM) sequences labeled in red color. B, Mutation type of different Wx edited lines. The Wx gene sequence at the targeted sites is shown. Wxb and Wxmp are used as controls. PAM sequences are in red. The mutated bases in different lines are marked as blue color with bold, and ‘-’ indicates nucleotides deletion. The mutated amino acid type and position is indicated in the brackets. C, Brown rice phenotypes of edited lines and their corresponding wild types. The Wxm4 mutant is milky white compared with other lines. SJ2, Songjing 2; LQD3, Longqingdao 3; SJ18, Suijing 18. Scale bars, 5 mm. D, Apparent amylose content in different mutation types compared with their corresponding wild type. Values are Mean ± SE (n = 3). P values were calculated by the Student’s t test (*, P < 0.05; **, P < 0.01).
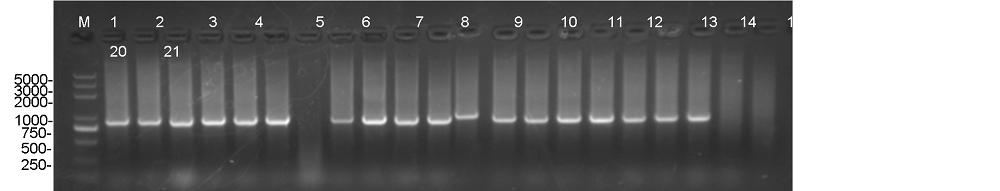
Supplemental Fig. 1. Identification of transgenic plants in T0 generation through PCR assay with hygromysin gene specific primer. M indicates DL2000 DNA Ladder Mix; Lines 1-19 are transgenic plants; Line 20 is WT, Line 21 is water used as a negative control.
| Primer name | Primer sequence (5'-3') |
|---|---|
| Wxb-sgRNA-F | CGACTCCACGCTTGTAGCAA |
| Wxb-sgRNA-R | TTGCTACAAGCGTGGAGTCG |
| Sequencing-Wxb-F | ATTGCTACAAGCGTGGAGTC |
| Sequencing-Wxb-R | CAGGGTAATCCTCGAAAGCG |
| HPT-F | TGCGCCCAAGCTGCATCAT |
| HPT-R | TGAACTCACCGCGACGTCTGT |
| Cas9T-F | AGCGGCAAGACTATCCTCGACT |
| Cas9T-R | TCAATCCTCTTCATGCGCTCCC |
Supplemental Table 1. Primers used in this study.
| Primer name | Primer sequence (5'-3') |
|---|---|
| Wxb-sgRNA-F | CGACTCCACGCTTGTAGCAA |
| Wxb-sgRNA-R | TTGCTACAAGCGTGGAGTCG |
| Sequencing-Wxb-F | ATTGCTACAAGCGTGGAGTC |
| Sequencing-Wxb-R | CAGGGTAATCCTCGAAAGCG |
| HPT-F | TGCGCCCAAGCTGCATCAT |
| HPT-R | TGAACTCACCGCGACGTCTGT |
| Cas9T-F | AGCGGCAAGACTATCCTCGACT |
| Cas9T-R | TCAATCCTCTTCATGCGCTCCC |
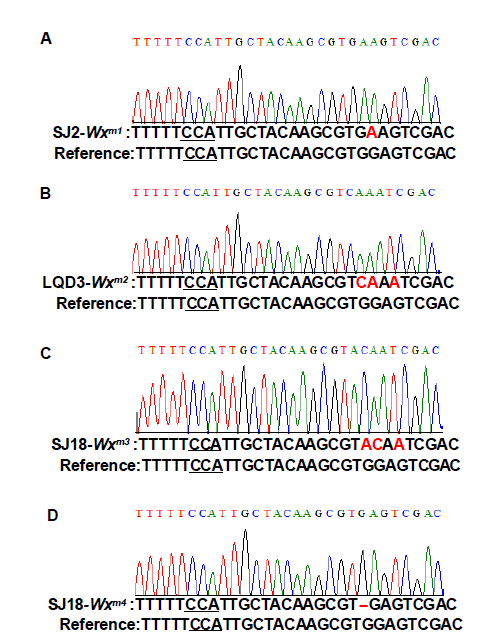
Supplemental Fig. 2. Identification of edited Wx mutants.Sequencing chromatograms and sequence of Wx genes in WT and different edited mutants are shown. The PAM (CCA) site is underlined and the edited bases are highlighted in red. ‘- ’ indicates nucleotides deletion.
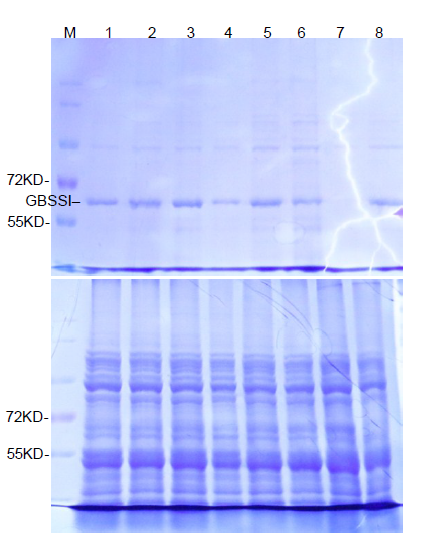
Supplemental Fig. 3. Detection of GBSSI in parents and edited lines. SDS-PAGE assay of starch granule-bound GBSSI (top) and total seed proteins (bottom) from mature seeds. M indicates protein marker; Line 1 is SJ2, Line 2 is Wxm1, Line 3 is LQD3, Line 4 is Wxm2, Line 5 is SJ18, Line 6 is Wxm3, Line 7 is Wxm4, Line 8 is Wxmq.
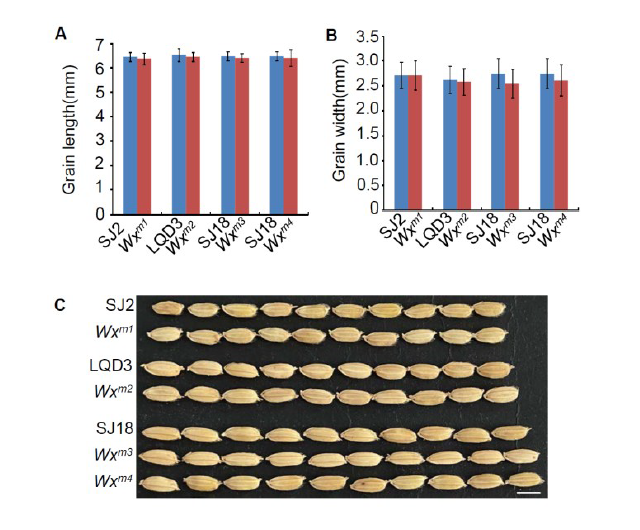
Supplemental Fig. 4. Grain morphology in parents and edited lines.(A and B) Quantification of grain length (A) and grain width (B) in WT and edited lines, respectively. Data are means±SE (n = 20).(C) The grain picture of WT and edited lines.
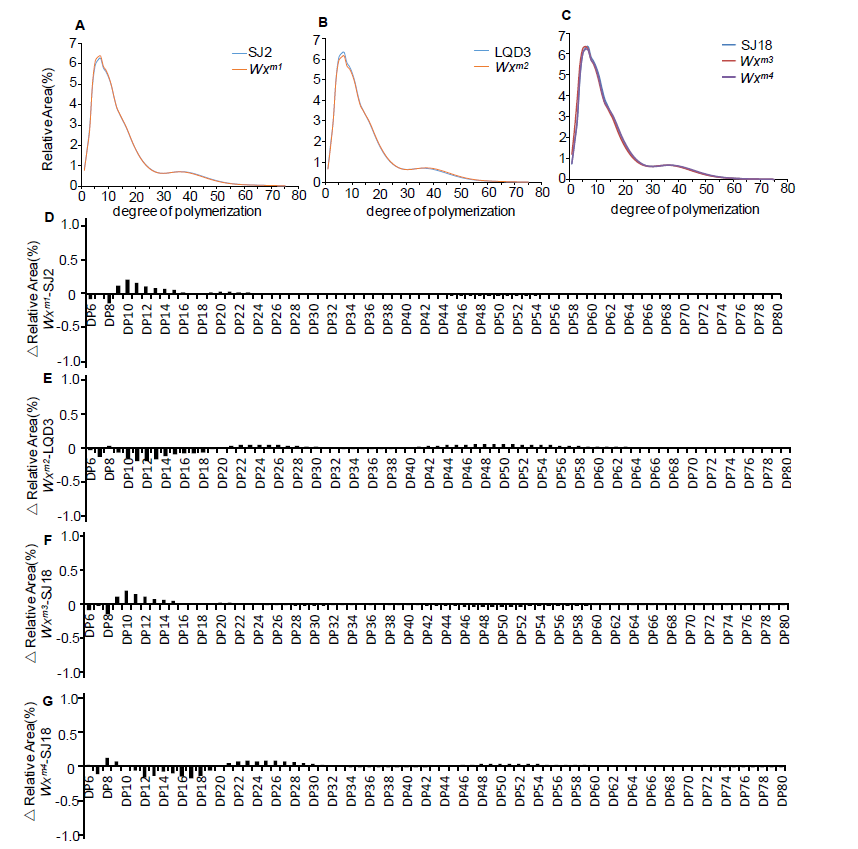
Supplemental Fig. 5. Amylopectin structure analysis in parents and edited lines.(A-C) indicate difference in chain length distribution of amylopectin in WT and edited lines. (D-G) show chain length fraction differences in WT and edited lines.
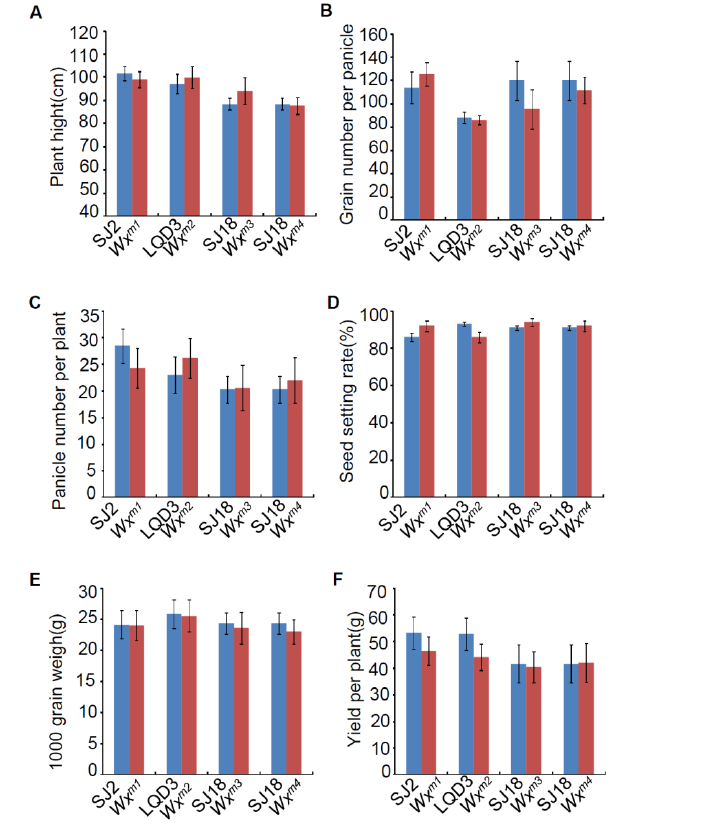
Supplemental Fig. 6. Agronomic traits analysis in parents and edited lines.After collected the plant, the above agronomic traits were investigated. For each mutation type, at least 10 individual plants were used for analysis. Data are means ± SE (n = 10).
| [1] | Fei Y Y, Yang J, Wang F Q, Fan F J, Li W Q, Wang J, Xu Y, Zhu J Y, Zhong W G.2019. Production of two elite glutinous rice varieties by editing Wx gene. Rice Sci, 26(2): 55-61. |
| [2] | Isshiki M, Morino K, Nakajima M, Okagaki R J, Wessler S R, Izawa T, Shimamoto K.1998. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J, 15(1): 133-138. |
| [3] | Juliano B O.1998. Varietal impact on rice quality. Cereal Foods World, 43(4): 207-222. |
| [4] | Khush G S.2005. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol, 59(1): 1-6. |
| [5] | Li M R, Li X X, Zhou Z J, Wu P Z, Fang M C, Pan X P, Lin Q P, Luo W B, Wu G J, Li H Q.2016. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci, 7: 377. |
| [6] | Li X F, Zhou W J, Ren Y K, Tian X J, Lv T X, Wang Z Y, Fang J, Chu C C, Yang J, Bu Q Y.2017. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J Genet Genom, 44: 175-178. |
| [7] | Liu Q Q, Wang Z Y, Chen X H, Cai X L, Tang S Z, Yu H X, Zhang J L, Hong M M, Gu M H.2003. Stable inheritance of the antisense Waxy gene in transgenic rice with reduced amylose level and improved quality. Transgenic Res, 12(1): 71-82. |
| [8] | Liu Q Q, Yu H X, Chen X H, Cai X L, Tang S Z, Wang Z Y, Gu M H.2005. Field performance of transgenic indica hybrid rice with improved cooking and eating quality by down-regulation of Wx gene expression. Mol Breeding, 16(3): 199-208. |
| [9] | Liu Q Q, Li Q F, Cai X L, Wang H M, Tang S Z, Yu H X, Wang Z Y, Gu M H.2006. Molecular marker-assisted selection for improved cooking and eating quality of two elite parents of hybrid rice. Acta Agron Sin, 46(6): 2354-2360. (in Chinese with English abstract) |
| [10] | Liu W Z, Xie X R, Ma X L, Li J, Chen J H, Liu Y G.2015. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant, 8(9): 1431-1433. |
| [11] | Ma X L, Zhang Q Y, Zhu Q Y, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G.2015. A robust CRISPR/Cas9 system for convenient, high- efficiency multiplex genome editing in monocot and dicot plants. Mol Plant, 8(8): 1274-1284. |
| [12] | Sato H, Suzuki Y, Sakai M, Imbe T.2002. Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice(Oryza sativa L.). Breeding Sci, 52: 131-135. |
| [13] | Sun Y W, Zhang X, Wu C Y, He Y B, Ma Y Z, Hou H, Guo X P, Du W M, Zhao Y D, Xia L Q.2016. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant, 9(4): 628-631. |
| [14] | Wang M G, Wang Z D, Mao Y F, Lu Y M, Yang R F, Tao X P, Zhu J K.2019. Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol J, 17(9): 1697-1699. |
| [15] | Wang Z Y, Zheng F Q, Shen G Z, Gao J P, Snustad D P, Li M G, Zhang J L, Hong M M.1995. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J, 7(4): 613-622. |
| [16] | Yao S, Chen T, Zhang Y D, Zhu Z, Zhao Q Y, Zhou L H, Zhao L, Zhao C F, Wang C L.2017. Pyramiding Pi-ta, Pi-b, and Wx-mq genes by marker-assisted selection in rice(Oryza sativa L.). Acta Agron Sin, 43(11): 1622-1631. (in Chinese with English abstract) |
| [17] | Yang J, Wang J, Fan F J, Zhu J Y, Chen T, Wang C L, Zheng T Q, Zhang J, Zhong W G, Xu J L.2013. Development of AS-PCR marker based on a key mutation confirmed by resequencing of Wx-mp in Milky Princess and its application in japonica soft rice(Oryza sativa L.) breeding. Plant Breeding, 132(6): 595-603. |
| [18] | Zhang J S, Zhang H, Botella J R, Zhu J K.2018. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol, 60(5): 369-375. |
| [19] | Zhou H, Xia D, He Y Q.2020. Rice grain quality-traditional traits for high quality rice and health-plus substances. Mol Breeding, 40(1): 1-17. |
| [20] | Zhu J H, Zhang C Q, Gu M H, Liu Q Q.2015. Progress in the allelic variation of Wx gene and its application in rice breeding. Chin J Rice Sci, 29(4): 431-438. (in Chinese with English abstract) |
| No related articles found! |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 1409310817
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 1201
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||